"Search peptides" will come in a future version. As fist step we will use this technology to better visualize peptide structures on the result page.
|
The Proteax
module of iScienceSearch provides an easy way to perform
substructure and structure similarity searches in major Internet
databases containing peptides, such as PubChem, CheBI and ChemSpider
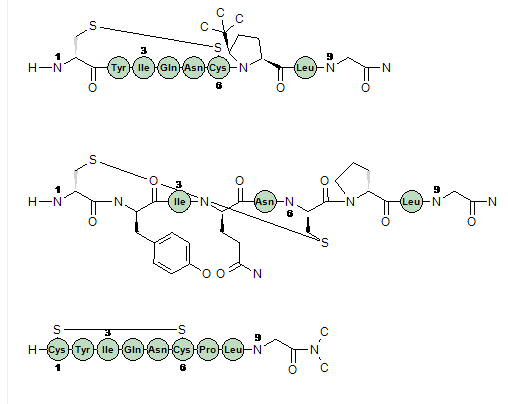
starting from plain 3-letter-code peptide sequences. When performing real structure searches, you can find peptides containing structure modified amino acid residues. These cannot be found using strictly text based searches such as BLAST. You can also search generic text terms in PubChem and CheBI and end up on a result page that only contain structures that are peptides or peptide like structures such as Penicillin. In order to analyze the results easily, we transform the complex structures back into the 3-letter-codes. We show structures when we cannot map back to the natural amino acids, see the picture below.
|